GGA

Galaxy Genome Annotation
View the Project on GitHub galaxy-genome-annotation/galaxy-genome-annotation.github.io
Galaxy Genome Annotation
GGA is a group of projects focused on supporting genome annotation inside Galaxy, in particular:
- Galaxy tools and workflows
- Training material
- Docker images for GMOD applications
- Python libraries to interact with GMOD applications
- Apollo tooling and service
Galaxy tools
Analysis tools
Various Galaxy tools for Genome Annotation are in the IUC repository, including: Augustus, BUSCO, FeelNC, Funannotate, Gffread, InterProScan, Maker, Metaeuk, Prokka, Repeat Masker, Repeat Modeler, Snap
Other tools are also available on other repositories: Antismash, Braker, EggNOG-Mapper
Visualisation tools
We are maintaining the following Galaxy tools for the visualisation of annotations:
- Circos: allows to build of a huge variety of highly configurable Circos plot.
- JBrowse: allows to generate fully customisable JBrowse instances.
- JBrowse2 (work in progress): based on the new generation of JBrowse.
Galaxy tools for GMOD apps
Galaxy tools to interact with GMOD applications (see below for the corresponding Docker images and Python libraries).
Training Material
Learn how to annotate new genomes by following the Genome Annotation tutorials on the Galaxy Training Network! Reusable workflows are provided together with detailed tutorials and videos.
Apollo tooling and service
Apollo server at UseGalaxy.eu
Based on the various components listed here, UseGalaxy.eu is hosting an Apollo server, freely accessible to the community.
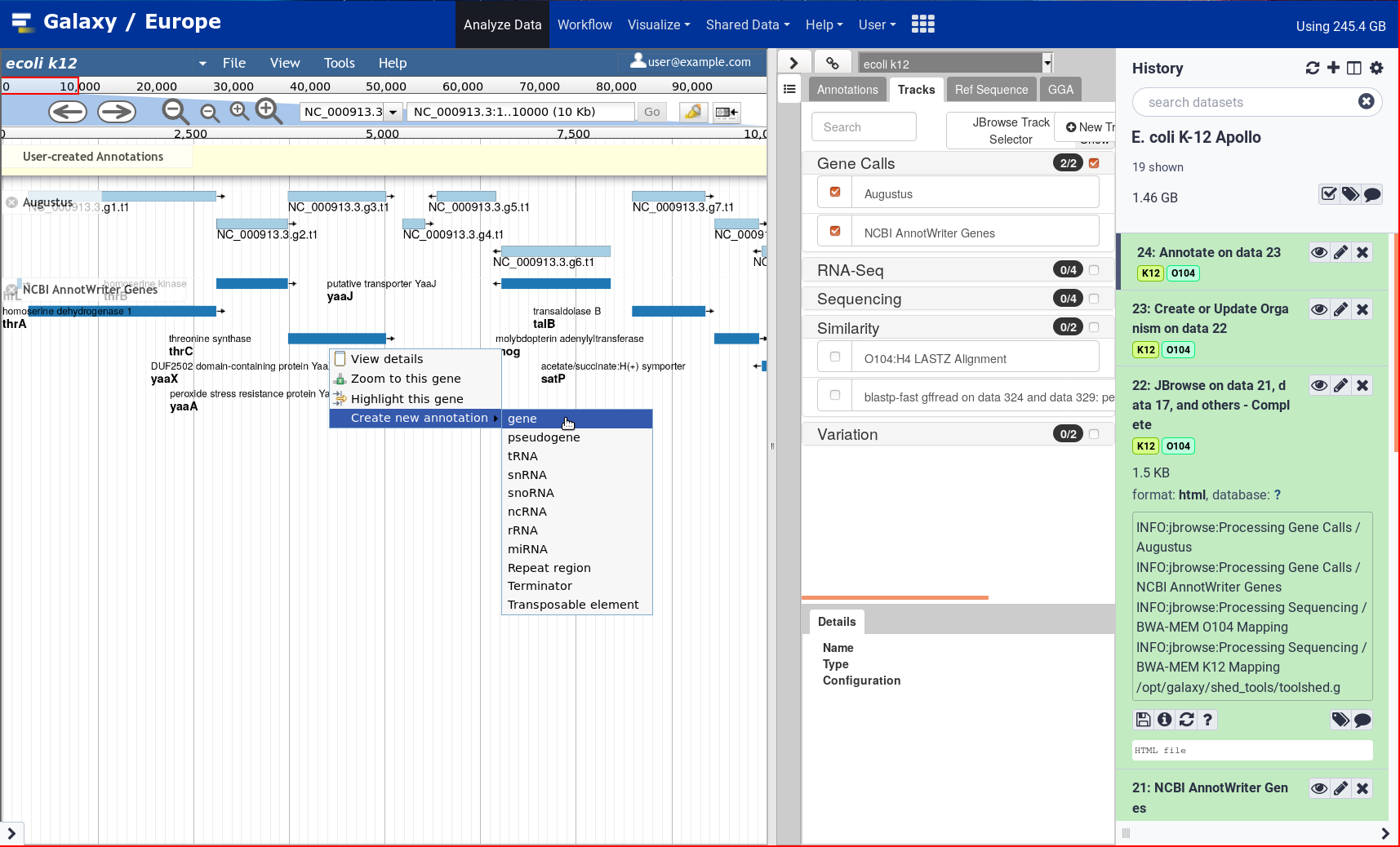
Using Galaxy, any user can upload its own organism, and start curating gene models collaboratively with other users.
Check out the GTN training material to learn how to use it!
Permapol
Permapol is a permission manager for Apollo, designed to ease collaboration when coupled with a Galaxy instance.
Apolpi
Apolpi is a tiny Flask application reimplementing a specific API function (organism listing) of Apollo, to make it run way faster.
Apollo report
Apollo report is a Docker image that contains everything needed to generate an Apollo per-user report each day. Based on the validation rules defined at http://bipaa.genouest.org/.
Docker images
docker-galaxy-genome-annotation
Galaxy Docker repository with tools for Genome Annotation. The image is built with tools for Assembly (Spades, Mira), Structural Prediction (Glimmer, Augustus), Functional Prediction (BLAST+, InterProScan, BLAST, Diamond, Blast2GO), various Utilities (FASTA manipulation tools, EMBOSS), tools for Comparative Genomics (CD-Hit, ClustalW, AntiSmash, mummer), and finally Annotation & Visualization tools (Apollo Tools, JBrowse-in-Galaxy, JBrowse-in-Galaxy Extras, Tripal Admin tools, Circos)
docker-tripal
A production ready Docker image for Tripal v2.x and v3.x.
docker-jbrowse
A production ready Docker image for JBrowse.
docker-chado
A production ready Docker image for Chado database. The image is built automatically using SQL dumps produced by chado-schema-builder, including up-to-date ontologies.
dockerized-gmod-deployment
If customizing the docker-galaxy-genome-annotation image isn’t your style, this is a preconfigured deployment of Galaxy + Apollo + Chado + Tripal + JBrowse + JBrowse REST API + PostGraphQL + JBrowse GraphQL Experiment all as a docker-compose.yml
Python libraries
python-apollo
Python library for talking to Apollo API. This includes the experimental Arrow Apollo client.
python-tripal
A Python library for interacting with Tripal
python-chado
A Python library for interacting with Chado database
Experiments
apollo-git-backup
Backup an apollo instance to a git repository
apollo-google-docs-integration
Experimental work for a plugin to allow referencing data from Apollo. For genomes which are re-opened / re-numbered / etc. between drafting and publication, it can be extremely helpful to reference persistent identifiers.
Support
This material is based upon work supported by the National Science Foundation under Grant Number (Award 1565146) and with the support of the Erasmus+ programme of the European Union (2020-1-NL01-KA203-064717).


























